By: Bradley Setzler
Re-posted from: http://juliaeconomics.com/2014/06/27/kalman-filter-for-panel-data-and-mle-in-julia-part-1/
* The script to reproduce the results of this tutorial in Julia is located here.
The Kalman Filter is notoriously difficult to estimate. I think this can be attributed to the following issues:
- It necessitates a large number of parameters that are difficult to manage.
- It requires the sequential inversion of matrices, so we must be careful to keep the matrices invertible and monitor numerical error.
- Most papers and Internet resources about the Kalman Filter assume that we have time-series rather than panel data, so they do not show how to adapt the parameters for panel data estimation
- Similarly, this literature makes “macro” assumptions like initializing at the steady state rather than the “micro” approach of estimating the initial state.
- Professional software is not well-suited to parameter estimation (more on this below).
In this tutorial, I will show how to write your own program that estimates the MLE of the most basic type of panel data Kalman Filter: time-invariant parameters and dedicated measures. It will explicitly impose assumptions to ensure identification, so that the estimates returned are uniquely determined from the data, unlike what would happen if we maximized the likelihood with the unrestricted Kalman Filter. In Part 2 of the tutorial, I generalize the estimation to permit a time-varying parameter space as well as time-varying parameter values, show how to identify the initial state, permit non-dedicated measures, and show how to “anchor” the model so that the units of the state space are determined to scale.
Kalman Filter DGP
The data generating process (DGP) corresponding to the panel data Kalman Filter is,
 ,
,
 ,
,
 ,
,
for time periods 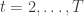 . We assume that
. We assume that 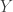 , referred to as “the measures” or “the data”, is the only observed term in this entire system. The first equation determines the evolution of the unobserved state,
, referred to as “the measures” or “the data”, is the only observed term in this entire system. The first equation determines the evolution of the unobserved state,  , and we will refer to it as the “transition equation”. The second equation determines the relationship between the state and measures or signals of the state,
, and we will refer to it as the “transition equation”. The second equation determines the relationship between the state and measures or signals of the state, 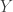 , and we will refer to it as the “measurement equation”. The shocks,
, and we will refer to it as the “measurement equation”. The shocks,  and
and 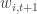 , are assumed to be independent across
, are assumed to be independent across 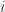 , across
, across  , and even across components so that
, and even across components so that 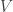 and
and  are diagonal.
are diagonal.
For our purposes, the parameters of interest are typically 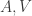 , although we generally need to identify
, although we generally need to identify 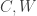 as well so that we can separate them from
as well so that we can separate them from 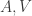 . It is no trouble to add other observables to these equations, e.g., in optimal control theory, there is usually an observed “input”, call it
. It is no trouble to add other observables to these equations, e.g., in optimal control theory, there is usually an observed “input”, call it  , on the right-hand side the transition equation, so that the equation takes the form,
, on the right-hand side the transition equation, so that the equation takes the form,
 ,
,
but this is a detail that could be added later with little difficulty, so we omit it until Part 2 of this tutorial. Also, we will assume for now that the initial distribution of the state is characterized by 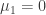 and
and 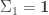 , but relax this assumption in Part 2.
, but relax this assumption in Part 2.
Failure of Identification and Necessary Assumptions
Before simulating the DGP, notice that it is indifferent to the scale or even the sign of  . In particular, consider dividing
. In particular, consider dividing  by a constant
by a constant  , and multiplying
, and multiplying 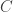 by
by  . Then,
. Then,
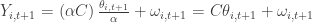 ,
,
so  cancels out and we are left with the same expression.
cancels out and we are left with the same expression.
Since our only information about  comes from
comes from 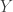 , this means that we will never be able to know
, this means that we will never be able to know 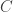 and
and  apart from one another, that is, we will never know the scale of
apart from one another, that is, we will never know the scale of  . Similarly, we cannot distinguish
. Similarly, we cannot distinguish  and
and  . By choosing
. By choosing 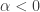 , we see that neither the sign nor the magnitude of
, we see that neither the sign nor the magnitude of  is determined by knowledge of
is determined by knowledge of 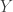 . Let me emphasize this point: we can change the sign of every entry in
. Let me emphasize this point: we can change the sign of every entry in  as we wish and get the same data. Since
as we wish and get the same data. Since  is one of the parameters of interest — in fact, our desire to learn
is one of the parameters of interest — in fact, our desire to learn  is often the motivation for using the Kalman Filter — this is a major issue. Since
is often the motivation for using the Kalman Filter — this is a major issue. Since  can take on a continuum of values, we see that the DGP results in the same
can take on a continuum of values, we see that the DGP results in the same 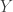 for a continuum of parameters
for a continuum of parameters  .
.
If we need to know the unique value of each parameter, we must make additional assumptions. The following assumptions, adapted from Cunha and Heckman (2008), are sufficient to ensure identification:
- Each of the measures (i.e., the components of
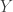 ) is a measure of only one of the factors (i.e., the components of
) is a measure of only one of the factors (i.e., the components of  ). This is the “dedicated measures” assumption, and it is equivalent to requiring each row of
). This is the “dedicated measures” assumption, and it is equivalent to requiring each row of 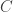 to have exactly one entry that is not assumed to be zero.
to have exactly one entry that is not assumed to be zero.
- There are at least 3 dedicated measures for each factor. This means that each column in
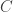 must have at least three entries that are not assumed to be zero.
must have at least three entries that are not assumed to be zero.
The first assumption can be relaxed a bit if there are more than 3 measures per factor, but we can never relax the assumption that there is at least one dedicated measure for one of the factors.
In practice, are these assumptions valid? Possibly, it depends on your faith in the measures available in your data. You must be able to confidently claim that certain measures in your data are measuring only one factor. The alternative is to abandon estimating the model. In other words, the Kalman Filter is not all-powerful — certain models simply cannot be estimated with certain data, and trying to force it to estimate a model that is not identified from your data will result in incorrect estimates.
Notice that professional Kalman Filter software does not allow one to impose these assumptions. This is one of the main motivations for this tutorial: at present, you must program the Kalman Filter yourself if you want valid estimates of the model parameters, and estimating the model parameters rather than projecting the state from given model parameters is the main purpose of the Kalman Filter in microeconomic applications.
Simulation of the Kalman Filter DGP
Before simulating the Kalman Filter DGP, we must first deal with the issue I mentioned at the beginning of this tutorial: the Kalman Filter DGP necessitates a large number of parameters that are difficult to manage. We need a function that can convert an array of parameters into the matrices 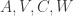 , including exponentiating variance terms so that when we use an optimizer later, the optimizer can try out different parameters without error. The following function, unpackParams, is my solution to this problem:
, including exponentiating variance terms so that when we use an optimizer later, the optimizer can try out different parameters without error. The following function, unpackParams, is my solution to this problem:
function unpackParams(params)
place = 0
A = reshape(params[(place+1):(place+stateDim^2)],(stateDim,stateDim))
place += stateDim^2
V = diagm(exp(params[(place+1):(place+stateDim)]))
place += stateDim
Cparams = params[(place+1):(place+stateDim*(obsDim-1))]
C = zeros(stateDim*obsDim,stateDim)
for j in [1:stateDim]
C[(1+obsDim*(j-1)):obsDim*j,j] = [1,Cparams[(1+(obsDim-1)*(j-1)):(obsDim-1)*j]]
end
place += (obsDim-1)*stateDim
W = diagm(exp(params[(place+1):(place+obsDim*stateDim)]))
return [&quot;A&quot;=&gt;A,&quot;V&quot;=&gt;V,&quot;C&quot;=&gt;C,&quot;W&quot;=&gt;W]
end
where stateDim is the length of  and obsDim is the number of measures on each component of
and obsDim is the number of measures on each component of  . This structure assumes that all measures are dedicated to a single factor, and the same number of measures is available for each factor. The function can be modified as needed to account for other structures. Here is the output from using this function on a set of parameters when there are two components of
. This structure assumes that all measures are dedicated to a single factor, and the same number of measures is available for each factor. The function can be modified as needed to account for other structures. Here is the output from using this function on a set of parameters when there are two components of  with 3 measures on each:
with 3 measures on each:
julia&gt; stateDims = 2
julia&gt; obsDims = 3
julia&gt; params0 = [1.,.3,.3,1.,0.,0.,.5,-.5,.5,-.5,0.,0.,0.,0.,0.,0.]
julia&gt; unpackParams(params0)
[&quot;V&quot;=&gt;2x2 Array{Float64,2}:
1.0 0.0
0.0 1.0,
&quot;C&quot;=&gt;6x2 Array{Float64,2}:
1.0 0.0
0.5 0.0
-0.5 0.0
0.0 1.0
0.0 0.5
0.0 -0.5,
&quot;A&quot;=&gt;2x2 Array{Float64,2}:
1.0 0.3
0.3 1.0,
&quot;W&quot;=&gt;6x6 Array{Float64,2}:
1.0 0.0 0.0 0.0 0.0 0.0
0.0 1.0 0.0 0.0 0.0 0.0
0.0 0.0 1.0 0.0 0.0 0.0
0.0 0.0 0.0 1.0 0.0 0.0
0.0 0.0 0.0 0.0 1.0 0.0
0.0 0.0 0.0 0.0 0.0 1.0]
Clearly, the most difficult part to construct is 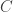 , the measurement matrix.
, the measurement matrix.
Now that we can convert an array of parameters into the appropriate parameter matrices, it is straight-forward to define a function that simulates the DGP of the Kalman Filter:
function KalmanDGP(params)
# initialize data
data = zeros(N,stateDim*obsDim*T)
# parameters
unpacked = unpackParams(params)
A = unpacked[&quot;A&quot;]
V = unpacked[&quot;V&quot;]
C = unpacked[&quot;C&quot;]
W = unpacked[&quot;W&quot;]
# draw from DGP
for i=1:N
# data of individual i
iData = ones(stateDim*obsDim*T)
current_state = rand(MvNormal(reshape(init_exp,(stateDim,)),init_var))
iData[1:stateDim*obsDim] = C*current_state + rand(MvNormal(W))
for t=2:T
current_state = A*current_state + rand(MvNormal(V))
iData[((t-1)*stateDim*obsDim+1):(t*stateDim*obsDim)] = C*current_state + rand(MvNormal(W))
end
# add individual to data
data[i,:] = iData
end
return data
end
Here is a simulated data set that satisfies the assumptions of time-invariant parameters and at least three dedicated measures (in particular, I use obsDims=4):
srand(2)
N = 1000
T = 4
stateDims = 2
obsDims = 4
init_exp = zeros(stateDim)
init_var = eye(stateDim)
data=KalmanDGP(params0)
Estimation Strategy for the Kalman Filter DGP
Now that we understand the Kalman Filter model, we can consider the Kalman Filter itself. The Kalman Filter is the estimator of what I have been calling the Kalman Filter DGP. The aim of the Kalman Filter for panel data is to estimate the mean and variance of  for each
for each 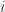 and each
and each  .
.
Estimation can be divided into two steps. Denoting 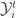 as the set of all data available at or before time
as the set of all data available at or before time  , the projection step is trivial to derive and given by:
, the projection step is trivial to derive and given by:
![\mathbb{E}\left[\theta_{i,t+1}\Big|\mathcal{Y}^t_i\right] = A\mathbb{E}\left[\theta_{i,t}\Big| \mathcal{Y}^t_i\right] \mathbb{E}\left[\theta_{i,t+1}\Big|\mathcal{Y}^t_i\right] = A\mathbb{E}\left[\theta_{i,t}\Big| \mathcal{Y}^t_i\right]](https://s0.wp.com/latex.php?latex=%5Cmathbb%7BE%7D%5Cleft%5B%5Ctheta_%7Bi%2Ct%2B1%7D%5CBig%7C%5Cmathcal%7BY%7D%5Et_i%5Cright%5D+%3D+A%5Cmathbb%7BE%7D%5Cleft%5B%5Ctheta_%7Bi%2Ct%7D%5CBig%7C+%5Cmathcal%7BY%7D%5Et_i%5Cright%5D&bg=ffffff&fg=333333&s=0)
![\mathbb{E}\left[Y_{i,t+1}\Big|\mathcal{Y}^t_i\right] = C\mathbb{E}\left[\theta_{i,t+1}\Big| \mathcal{Y}^t_i\right] \mathbb{E}\left[Y_{i,t+1}\Big|\mathcal{Y}^t_i\right] = C\mathbb{E}\left[\theta_{i,t+1}\Big| \mathcal{Y}^t_i\right]](https://s0.wp.com/latex.php?latex=%5Cmathbb%7BE%7D%5Cleft%5BY_%7Bi%2Ct%2B1%7D%5CBig%7C%5Cmathcal%7BY%7D%5Et_i%5Cright%5D+%3D+C%5Cmathbb%7BE%7D%5Cleft%5B%5Ctheta_%7Bi%2Ct%2B1%7D%5CBig%7C+%5Cmathcal%7BY%7D%5Et_i%5Cright%5D&bg=ffffff&fg=333333&s=0)
![\mathrm{Var}\left[Y_{i,t+1}\Big|\mathcal{Y}^t_i\right] = A\mathbb{E}\left[\theta_{i,t}\Big| \mathcal{Y}^t_i\right]A'+V \mathrm{Var}\left[Y_{i,t+1}\Big|\mathcal{Y}^t_i\right] = A\mathbb{E}\left[\theta_{i,t}\Big| \mathcal{Y}^t_i\right]A'+V](https://s0.wp.com/latex.php?latex=%5Cmathrm%7BVar%7D%5Cleft%5BY_%7Bi%2Ct%2B1%7D%5CBig%7C%5Cmathcal%7BY%7D%5Et_i%5Cright%5D+%3D+A%5Cmathbb%7BE%7D%5Cleft%5B%5Ctheta_%7Bi%2Ct%7D%5CBig%7C+%5Cmathcal%7BY%7D%5Et_i%5Cright%5DA%27%2BV&bg=ffffff&fg=333333&s=0)
![\mathrm{Var}\left[\theta_{i,t+1}\Big|\mathcal{Y}^t_i\right] = C\mathbb{E}\left[\theta_{i,t}\Big| \mathcal{Y}^t_i\right]C'+W \mathrm{Var}\left[\theta_{i,t+1}\Big|\mathcal{Y}^t_i\right] = C\mathbb{E}\left[\theta_{i,t}\Big| \mathcal{Y}^t_i\right]C'+W](https://s0.wp.com/latex.php?latex=%5Cmathrm%7BVar%7D%5Cleft%5B%5Ctheta_%7Bi%2Ct%2B1%7D%5CBig%7C%5Cmathcal%7BY%7D%5Et_i%5Cright%5D+%3D+C%5Cmathbb%7BE%7D%5Cleft%5B%5Ctheta_%7Bi%2Ct%7D%5CBig%7C+%5Cmathcal%7BY%7D%5Et_i%5Cright%5DC%27%2BW&bg=ffffff&fg=333333&s=0)
The update step is:
![\mathbb{E}\left[\theta_{i,t+1}\Big|\mathcal{Y}^{t+1}_i\right] = \mathbb{E}\left[\theta_{i,t+1}\Big| \mathcal{Y}^t_i\right]+K_{t+1} \tilde{Y}_{i,t+1} \mathbb{E}\left[\theta_{i,t+1}\Big|\mathcal{Y}^{t+1}_i\right] = \mathbb{E}\left[\theta_{i,t+1}\Big| \mathcal{Y}^t_i\right]+K_{t+1} \tilde{Y}_{i,t+1}](https://s0.wp.com/latex.php?latex=%5Cmathbb%7BE%7D%5Cleft%5B%5Ctheta_%7Bi%2Ct%2B1%7D%5CBig%7C%5Cmathcal%7BY%7D%5E%7Bt%2B1%7D_i%5Cright%5D+%3D+%5Cmathbb%7BE%7D%5Cleft%5B%5Ctheta_%7Bi%2Ct%2B1%7D%5CBig%7C+%5Cmathcal%7BY%7D%5Et_i%5Cright%5D%2BK_%7Bt%2B1%7D+%5Ctilde%7BY%7D_%7Bi%2Ct%2B1%7D&bg=ffffff&fg=333333&s=0)
![\mathrm{Var}\left[\theta_{i,t+1}\Big|\mathcal{Y}^{t+1}_i\right] = \mathrm{Var}\left[\theta_{i,t+1}\Big| \mathcal{Y}^{t}_i\right]-K_{t+1} C\mathrm{Var}\left[\theta_{i,t+1}\Big| \mathcal{Y}^{t}_i\right] \mathrm{Var}\left[\theta_{i,t+1}\Big|\mathcal{Y}^{t+1}_i\right] = \mathrm{Var}\left[\theta_{i,t+1}\Big| \mathcal{Y}^{t}_i\right]-K_{t+1} C\mathrm{Var}\left[\theta_{i,t+1}\Big| \mathcal{Y}^{t}_i\right]](https://s0.wp.com/latex.php?latex=%5Cmathrm%7BVar%7D%5Cleft%5B%5Ctheta_%7Bi%2Ct%2B1%7D%5CBig%7C%5Cmathcal%7BY%7D%5E%7Bt%2B1%7D_i%5Cright%5D+%3D+%5Cmathrm%7BVar%7D%5Cleft%5B%5Ctheta_%7Bi%2Ct%2B1%7D%5CBig%7C+%5Cmathcal%7BY%7D%5E%7Bt%7D_i%5Cright%5D-K_%7Bt%2B1%7D+C%5Cmathrm%7BVar%7D%5Cleft%5B%5Ctheta_%7Bi%2Ct%2B1%7D%5CBig%7C+%5Cmathcal%7BY%7D%5E%7Bt%7D_i%5Cright%5D&bg=ffffff&fg=333333&s=0)
where,
![\tilde{Y}_{t+1}=Y_{t+1}-\mathbb{E}\left[Y_{i,t+1}\Big|\mathcal{Y}^{t}_i\right] \tilde{Y}_{t+1}=Y_{t+1}-\mathbb{E}\left[Y_{i,t+1}\Big|\mathcal{Y}^{t}_i\right]](https://s0.wp.com/latex.php?latex=%5Ctilde%7BY%7D_%7Bt%2B1%7D%3DY_%7Bt%2B1%7D-%5Cmathbb%7BE%7D%5Cleft%5BY_%7Bi%2Ct%2B1%7D%5CBig%7C%5Cmathcal%7BY%7D%5E%7Bt%7D_i%5Cright%5D&bg=ffffff&fg=333333&s=0)
is the error from projecting 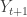 at time
at time  , and,
, and,
![K_{t+1}=\mathrm{Var}\left[\theta_{i,t+1}\Big|\mathcal{Y}^{t}_i\right]C'\left(\mathrm{Var}\left[Y_{i,t+1}\Big|\mathcal{Y}^{t}_i\right]\right)^{-1} K_{t+1}=\mathrm{Var}\left[\theta_{i,t+1}\Big|\mathcal{Y}^{t}_i\right]C'\left(\mathrm{Var}\left[Y_{i,t+1}\Big|\mathcal{Y}^{t}_i\right]\right)^{-1}](https://s0.wp.com/latex.php?latex=K_%7Bt%2B1%7D%3D%5Cmathrm%7BVar%7D%5Cleft%5B%5Ctheta_%7Bi%2Ct%2B1%7D%5CBig%7C%5Cmathcal%7BY%7D%5E%7Bt%7D_i%5Cright%5DC%27%5Cleft%28%5Cmathrm%7BVar%7D%5Cleft%5BY_%7Bi%2Ct%2B1%7D%5CBig%7C%5Cmathcal%7BY%7D%5E%7Bt%7D_i%5Cright%5D%5Cright%29%5E%7B-1%7D&bg=ffffff&fg=333333&s=0)
is called the optimal Kalman gain and is derived as the solution to a least-squares problem.
The individual likelihood of the Kalman Filter is given by,
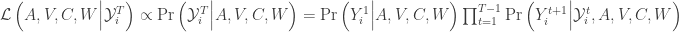
The initial conditions enter through  . In this tutorial, we assume
. In this tutorial, we assume  , where
, where 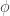 is the multivariate normal PDF. We permit individual-specific initial conditions and unknown parameters in Part 2 of the tutorial. Furthermore,
is the multivariate normal PDF. We permit individual-specific initial conditions and unknown parameters in Part 2 of the tutorial. Furthermore, ![\Pr\left(Y^{t+1}_i\Big|\mathcal{Y}^t_i,A,V,C,W\right)=\phi\left(Y_i^{t+1}\big|\mathbb{E}\left[Y_{i,t+1}\Big|\mathcal{Y}^t_i\right],\mathrm{Var}\left[Y_{i,t+1}\Big|\mathcal{Y}^t_i\right]\right) \Pr\left(Y^{t+1}_i\Big|\mathcal{Y}^t_i,A,V,C,W\right)=\phi\left(Y_i^{t+1}\big|\mathbb{E}\left[Y_{i,t+1}\Big|\mathcal{Y}^t_i\right],\mathrm{Var}\left[Y_{i,t+1}\Big|\mathcal{Y}^t_i\right]\right)](https://s0.wp.com/latex.php?latex=%5CPr%5Cleft%28Y%5E%7Bt%2B1%7D_i%5CBig%7C%5Cmathcal%7BY%7D%5Et_i%2CA%2CV%2CC%2CW%5Cright%29%3D%5Cphi%5Cleft%28Y_i%5E%7Bt%2B1%7D%5Cbig%7C%5Cmathbb%7BE%7D%5Cleft%5BY_%7Bi%2Ct%2B1%7D%5CBig%7C%5Cmathcal%7BY%7D%5Et_i%5Cright%5D%2C%5Cmathrm%7BVar%7D%5Cleft%5BY_%7Bi%2Ct%2B1%7D%5CBig%7C%5Cmathcal%7BY%7D%5Et_i%5Cright%5D%5Cright%29&bg=ffffff&fg=333333&s=0) .
.
Finally, since individuals are assumed to be independent,

Now that we have a simple expression for the likelihood, we need only convert it into a Julia code.
Estimation of the Kalman Filter in Julia
To simplify computation, we write a single function that simultaneously predicts and updates (in that order) from any time period  to the next time period
to the next time period 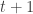 . It begins with the “posterior” estimates of the expected state and variance of the state at time
. It begins with the “posterior” estimates of the expected state and variance of the state at time  , that is,
, that is, ![\mathbb{E}\left[\theta_{i,t}\Big|\mathcal{Y}^{t}_i\right],\mathrm{Var}\left[\theta_{i,t}\Big|\mathcal{Y}^{t}_i\right] \mathbb{E}\left[\theta_{i,t}\Big|\mathcal{Y}^{t}_i\right],\mathrm{Var}\left[\theta_{i,t}\Big|\mathcal{Y}^{t}_i\right]](https://s0.wp.com/latex.php?latex=%5Cmathbb%7BE%7D%5Cleft%5B%5Ctheta_%7Bi%2Ct%7D%5CBig%7C%5Cmathcal%7BY%7D%5E%7Bt%7D_i%5Cright%5D%2C%5Cmathrm%7BVar%7D%5Cleft%5B%5Ctheta_%7Bi%2Ct%7D%5CBig%7C%5Cmathcal%7BY%7D%5E%7Bt%7D_i%5Cright%5D&bg=ffffff&fg=333333&s=0) , uses these to predict the “prior” estimates at time
, uses these to predict the “prior” estimates at time 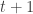 , that is,
, that is, ![\mathbb{E}\left[\theta_{i,t+1}\Big|\mathcal{Y}^{t}_i\right],\mathrm{Var}\left[\theta_{i,t+1}\Big|\mathcal{Y}^{t}_i\right] \mathbb{E}\left[\theta_{i,t+1}\Big|\mathcal{Y}^{t}_i\right],\mathrm{Var}\left[\theta_{i,t+1}\Big|\mathcal{Y}^{t}_i\right]](https://s0.wp.com/latex.php?latex=%5Cmathbb%7BE%7D%5Cleft%5B%5Ctheta_%7Bi%2Ct%2B1%7D%5CBig%7C%5Cmathcal%7BY%7D%5E%7Bt%7D_i%5Cright%5D%2C%5Cmathrm%7BVar%7D%5Cleft%5B%5Ctheta_%7Bi%2Ct%2B1%7D%5CBig%7C%5Cmathcal%7BY%7D%5E%7Bt%7D_i%5Cright%5D&bg=ffffff&fg=333333&s=0) , uses the priors to obtain the likelihood of the new observation at time
, uses the priors to obtain the likelihood of the new observation at time 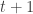 , and finally uses the priors as well as the new observation at time
, and finally uses the priors as well as the new observation at time 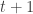 to update to the posterior estimates at time
to update to the posterior estimates at time 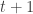 . The function, incrementKF (“a time increment of the Kalman Filter”, is as follows:
. The function, incrementKF (“a time increment of the Kalman Filter”, is as follows:
function incrementKF(params,post_exp,post_var,new_obs)
# unpack parameters
unpacked = unpackParams(params)
A = unpacked[&quot;A&quot;]
V = unpacked[&quot;V&quot;]
C = unpacked[&quot;C&quot;]
W = unpacked[&quot;W&quot;]
# predict
prior_exp = A*post_exp
prior_var = A*post_var*A' + V
obs_prior_exp = C*prior_exp
obs_prior_var = C*prior_var*C' + W
# update
residual = new_obs - obs_prior_exp
obs_prior_cov = prior_var*C'
kalman_gain = obs_prior_cov*inv(obs_prior_var)
post_exp = prior_exp + kalman_gain*residual
post_var = prior_var - kalman_gain*obs_prior_cov'
# step likelihood
dist = MvNormal(reshape(obs_prior_exp,(length(obs_prior_exp),)),obs_prior_var)
log_like = logpdf(dist,new_obs)
return [&quot;post_exp&quot;=&gt;post_exp,&quot;post_var&quot;=&gt;post_var,&quot;log_like&quot;=&gt;log_like]
end
We see that the code is quite simple, as it closely mimics the mathematical formulas above in syntax. The part of the code that does not mimic the usual mathematical syntax — preparing parameters — is handled elsewhere by unpackParams.
Now that we have the code that predicts and updates the Kalman Filter for any individual while outputting the contribution to the log-likelihood, we can iterate over time and collect all of the log-likelihood contributions from an individual’s time path:
function indivKF(params,init_exp,init_var,i)
iData = data[i,:]
# initialization
post_exp = init_exp
post_var = init_var
init_obs = matrix([iData[obsDict[1]]])'
dist = MvNormal(eye(length(init_obs)))
log_like=logpdf(dist,init_obs)
for t = 1:(T-1)
# predict and update
new_obs = matrix([iData[obsDict[t+1]]])'
new_post = incrementKF(params,post_exp,post_var,new_obs)
# replace
post_exp = new_post[&quot;post_exp&quot;]
post_var = new_post[&quot;post_var&quot;]
# contribute
log_like += new_post[&quot;log_like&quot;]
end
return log_like
end
This initializes the individual’s contribution to the likelihood using the assumed initial distribution of  , then proceeds to predict and update the Kalman Filter over time until all log-likelihood contributions have been collected. The only confusing part is obsDict, which is a dictionary containing the column names of the observations, which are assumed to be stored in a DataFrame called data. This strategy isn’t necessary, but it avoids a lot of complications, and also makes our example better resemble a real-world application to a data set.
, then proceeds to predict and update the Kalman Filter over time until all log-likelihood contributions have been collected. The only confusing part is obsDict, which is a dictionary containing the column names of the observations, which are assumed to be stored in a DataFrame called data. This strategy isn’t necessary, but it avoids a lot of complications, and also makes our example better resemble a real-world application to a data set.
Finally, we loop through each individual, summing all of their lifetime log-likelihood contributions:
function sampleKF(params,init_exp,init_var)
log_like = 0.0
N = size(data,1)
for i in 1:N
log_like += indivKF(params,init_exp,init_var,i)
end
neg_avg_log_like = -log_like/N
println(&quot;current average negative log-likelihood: &quot;,neg_avg_log_like)
return neg_avg_log_like[1]
end
where I like to include a print statement so that I can monitor the convergence of the likelihood. Thus, we have the panel data Kalman Filter likelihood, under the appropriate assumptions to ensure identification. Finally, we use a wrapper to prepare it for optimization:
function wrapLoglike(params)
print(&quot;current parameters: &quot;,params)
return sampleKF(params,init_exp,init_var)
end
where I have included a print statement so that I can monitor the parameters that the optimizer is testing and know in advance if the estimates are diverging (if it were diverging, I would know my code has a bug and could stop the optimizer before it wastes too much time).
We conclude by estimating the Kalman Filter MLE on simulated data.
Results
First, we simulate data from KalmanDGP:
srand(2)
N = 1000
T = 4
stateDim = 2
obsDim = 3
init_exp = zeros(stateDim)
init_var = eye(stateDim)
params0 = [1.,0.,0.,1.,0.,0.,.5,-.5,.5,-.5,0.,0.,0.,0.,0.,0.]
data=KalmanDGP(params0)
data = DataFrame(data)
colnames!(data,[&quot;one_1&quot;,&quot;one_2&quot;,&quot;one_3&quot;,&quot;one_4&quot;,&quot;one_5&quot;,&quot;one_6&quot;,&quot;two_1&quot;,&quot;two_2&quot;,&quot;two_3&quot;,&quot;two_4&quot;,&quot;two_5&quot;,&quot;two_6&quot;,&quot;three_1&quot;,&quot;three_2&quot;,&quot;three_3&quot;,&quot;three_4&quot;,&quot;three_5&quot;,&quot;three_6&quot;,&quot;four_1&quot;,&quot;four_2&quot;,&quot;four_3&quot;,&quot;four_4&quot;,&quot;four_5&quot;,&quot;four_6&quot;,&quot;outcome&quot;])
obsDict = {[&quot;one_1&quot;,&quot;one_2&quot;,&quot;one_3&quot;,&quot;one_4&quot;,&quot;one_5&quot;,&quot;one_6&quot;],[&quot;two_1&quot;,&quot;two_2&quot;,&quot;two_3&quot;,&quot;two_4&quot;,&quot;two_5&quot;,&quot;two_6&quot;],[&quot;three_1&quot;,&quot;three_2&quot;,&quot;three_3&quot;,&quot;three_4&quot;,&quot;three_5&quot;,&quot;three_6&quot;],[&quot;four_1&quot;,&quot;four_2&quot;,&quot;four_3&quot;,&quot;four_4&quot;,&quot;four_5&quot;,&quot;four_6&quot;]}
Everything here is the same as in the DGP code above, except that we are converting the data matrix into a DataFrame. The columns are named using the syntax time_number, so that three_4 means the 4th observation in time period 3. It creates obsDict such that, for example, data[obsDict[3]] would return the variables from three_1 to three_6 – all of the observations at time 3 in the correct order.
Now, we are ready to estimate the Kalman Filter as follows:
MLE = optimize(wrapLoglike,params0,method=:cg,ftol=1e-8)
When it is finished (and we have watched it converge using the print statements), we can view the parameters in the appropriate context by:
julia&gt; unpackParams(MLE.minimum)
[&quot;V&quot;=&gt;2x2 Array{Float64,2}:
1.11323 0.0
0.0 0.918925,
&quot;C&quot;=&gt;6x2 Array{Float64,2}:
1.0 0.0
0.489902 0.0
-0.490378 0.0
0.0 1.0
0.0 0.528178
0.0 -0.517231,
&quot;A&quot;=&gt;2x2 Array{Float64,2}:
0.976649 -0.00976326
0.00762499 0.994169 ,
&quot;W&quot;=&gt;6x6 Array{Float64,2}:
0.977998 0.0 0.0 0.0 0.0 0.0
0.0 0.97972 0.0 0.0 0.0 0.0
0.0 0.0 1.0241 0.0 0.0 0.0
0.0 0.0 0.0 0.990665 0.0 0.0
0.0 0.0 0.0 0.0 1.00625 0.0
0.0 0.0 0.0 0.0 0.0 1.01042]
This compares favorably to the true parameters,
julia&gt; unpackParams(params0)
[&quot;V&quot;=&gt;2x2 Array{Float64,2}:
1.0 0.0
0.0 1.0,
&quot;C&quot;=&gt;6x2 Array{Float64,2}:
1.0 0.0
0.5 0.0
-0.5 0.0
0.0 1.0
0.0 0.5
0.0 -0.5,
&quot;A&quot;=&gt;2x2 Array{Float64,2}:
1.0 0.0
0.0 1.0,
&quot;W&quot;=&gt;6x6 Array{Float64,2}:
1.0 0.0 0.0 0.0 0.0 0.0
0.0 1.0 0.0 0.0 0.0 0.0
0.0 0.0 1.0 0.0 0.0 0.0
0.0 0.0 0.0 1.0 0.0 0.0
0.0 0.0 0.0 0.0 1.0 0.0
0.0 0.0 0.0 0.0 0.0 1.0]
On a single processor of my personal laptop, optimization requires 446 seconds. In response to a reader comment, I also added this version of the code that achieves optimization in 393 seconds by removing all global variables.
Bradley J. Setzler

